Beyond Myocytes: Benchmarking Microstructural Heterogeneity in Cardiac Electrophysiology
- chair:Computational Cardiac Modeling
- type:Master thesis
- tutor:
Beyond Myocytes: Benchmarking Microstructural Heterogeneity in Cardiac Electrophysiology
Introduction. Heart muscle contraction is determined by excitation waves travelling through its tissue, where traditionally, these waves are viewed to be transmitted only via electrically coupled excitable cardiomyocytes (CM). However, increasing evidence shows that also non-myocytes (NM), e.g. fibroblasts, are electrically coupled to CM and may thus modulate excitation spread [1].
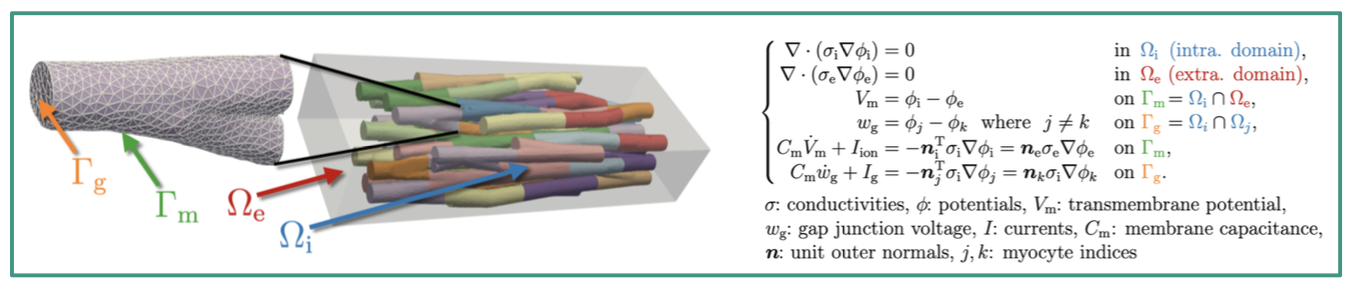
Left: An example mesh suitable for EMI model simulations, containing explicitly represented myocytes (shown in different colors and summarized into intracellular domain Ωi) surrounded by an extracellular bath, Ωe, shown in grey. Generated with [2], illustrated by Joshua Steyer.
Right: EMI model equations describing the spatiotemporal behaviour of excitation via the transmembrane voltage, Vm.
Thesis Topic. We are working with the novel extracellular-membrane-intracellular (EMI) model [3] (see the upper Figure), which explicitly resolves CM and the surrounding extracellular space within the computational mesh to study the micro-structural behaviour of excitation spread in cardiac tissue. So far, simulations with the EMI model have only considered CM and a homogenous extracellular space. The aim of this thesis is thus, to use the electrophysiology simulator openCARP [4] to carry out simulations on a suite of predefined benchmark problems covering NM and a heterogeneous extracellular space composition (e.g. to consider collagen presence). In addition to designing the geometric representation of different cell types in the discretised space (mesh), we want to employ different ionic models suitable to model the membrane dynamics and thus, the electrophysiological behaviour of different cell types.
Your tasks will be to:
- Create a suite of reproducible benchmark configurations based on structured meshes, in which box-like regions are defined to represent different cell types, sizes and alignments
- Define and investigate research questions within the benchmark configurations, based on experimental and/or in silico results of NM-mediated conduction in the heart (e.g. a test of the previously assumed but unproven passive conduction via NM or saltatory conduction via CM in NM-rich tissue)
- Carry out corresponding simulations in the openCARP electrophysiology simulator and critically
discuss and contextualise the outcomes
Further software and tools used (advantageous, but not required):
- ParaView or Meshalyzer for visualisation
- Python for mesh generation as well as pre- and postprocessing of simulations
- bwUniCluster3.0 to carry out simulations
- Computational cardiac electrophysiology (Courtemanche, Sachse and Plonsey models)
Sources
[1] Giardini et al. Correlative imaging integrates electrophysiology with three-dimensional murine heart reconstruction to reveal electrical coupling between cell types, Nat Cardiovasc Res (2025), doi: 10.1038/s44161-025-00728-9
[2] Potse M, Cirrottola L, Froehly A. A practical algorithm to build geometric models of cardiac muscle structure. In: 8th ECCOMAS. Oslo, Norway (2022)
[3] Tveito et al. Modeling Excitable Tissue: The EMI Framework. Simula Springer Briefs on Computing. Springer International Publishing (2020)
[4] https://opencarp.org/

