Simulation of 12-lead surface ECGs to classify cardiovascular diseases using machine learning techniques
- contact:
The electrocardiogram (ECG) is a non-invasive and cost-effective tool for the initial examination and monitoring of patients presenting with cardiac complaints, such as among others atrial flutter or ischemia. The rising number of patients suffering from cardiovascular diseases worldwide together with future telemedicine and home monitoring systems will boost the need for automated and validated ECG analysis.
Recently, machine learning (ML) techniques have been proposed for the detection and classification of cardiac diseases based on ECG traces. However, key challenges of ML are the investigation of the influence of data uncertainty and the assessment of the techniques’ uncertainty itself. Hence, there is a strong need for a metrological validation of ECG analysis algorithms using reference data with a traceable ground truth. Ground truth in medicine is a challenge and is usually addressed either by consensus of multiple experts or by using synthetic data.
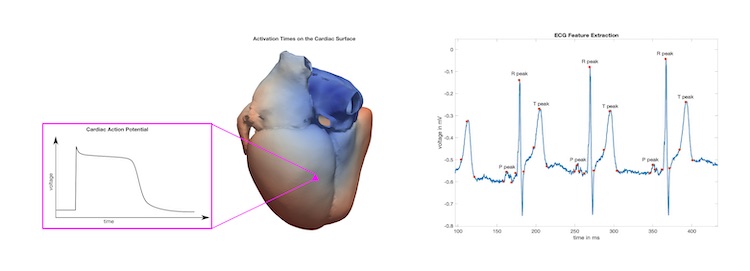
Therefore, the aim of this project is to develop a large synthetic ECG database for the uncertainty quantification and benchmarking of different analysis algorithms. For this purpose, 10.000 12-lead surface ECG traces will be simulated using an electrophysiological modeling framework. The simulated ECGs are approved regarding their closeness to reality if a physician can't distinguish between the in silico and real clinical recordings in a blinded experiment. In doing so, an ECG reference database of a representative virtual population that includes healthy variation as well as selected pathologies will be generated and tagged with its respective labels of the modeled cardiac disease.