Computer simulation to relate patterns of fibrosis to arrhythmogenicity in atrial fibrillation
- contact:
- project group:
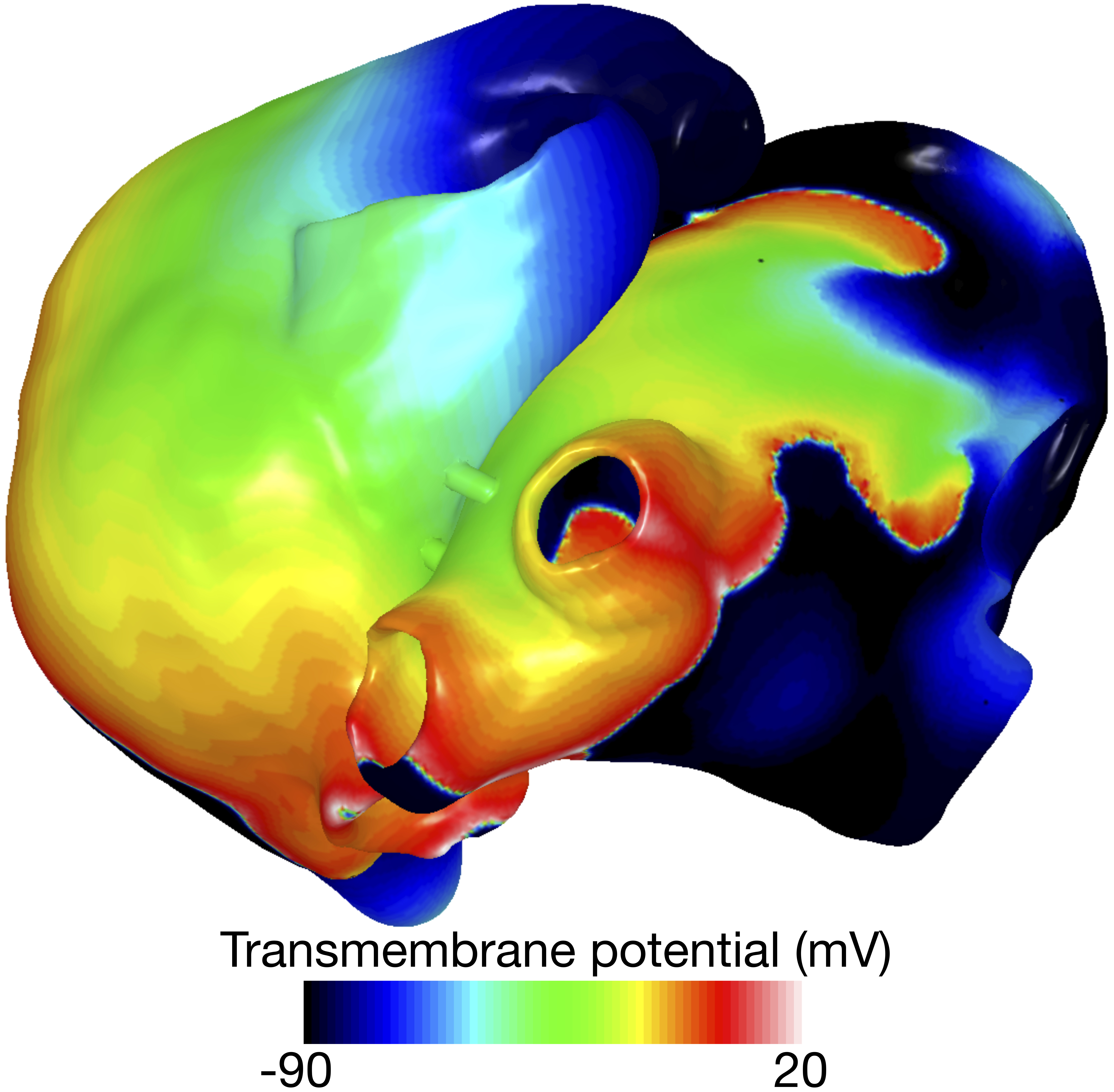
There are several methods to simulate cardiac electrophysiology at tissue level. The biophysical models governed by the bidomain and monodomain equations are the most commonly used models. These models can reproduce complex mechanisms leading to reentry and atrial fibrillation. In the figure below, you can see Luca Azzolin’s results, in another project of our team, using the monodomain equation to generate reentry. However, the high computational cost renders these approaches unsuitable for numerous clinical settings. On the other hand, models based on the Eikonal equation are an alternative that require significantly shorter simulations times. Unfortunately, Eikonal models are still incapable of reproducing phenomena such as unidirectional block and reentry in heterogeneous and anisotropic tissue.
The first goal of this project is to develop an upgraded Eikonal model capable of reproducing functional reentry. Our results will allow fast simulations of atrial fibrillation patients with heterogenous fibrosis. Once the model is implemented, the second goal will be to assess different patterns of fibrosis and their arrhythmogenicity. Initially relatively simple patterns will be analyzed. Eventually, their complexity will increase towards the end of the project.